Connect with us
Published
2 weeks agoon
By
admin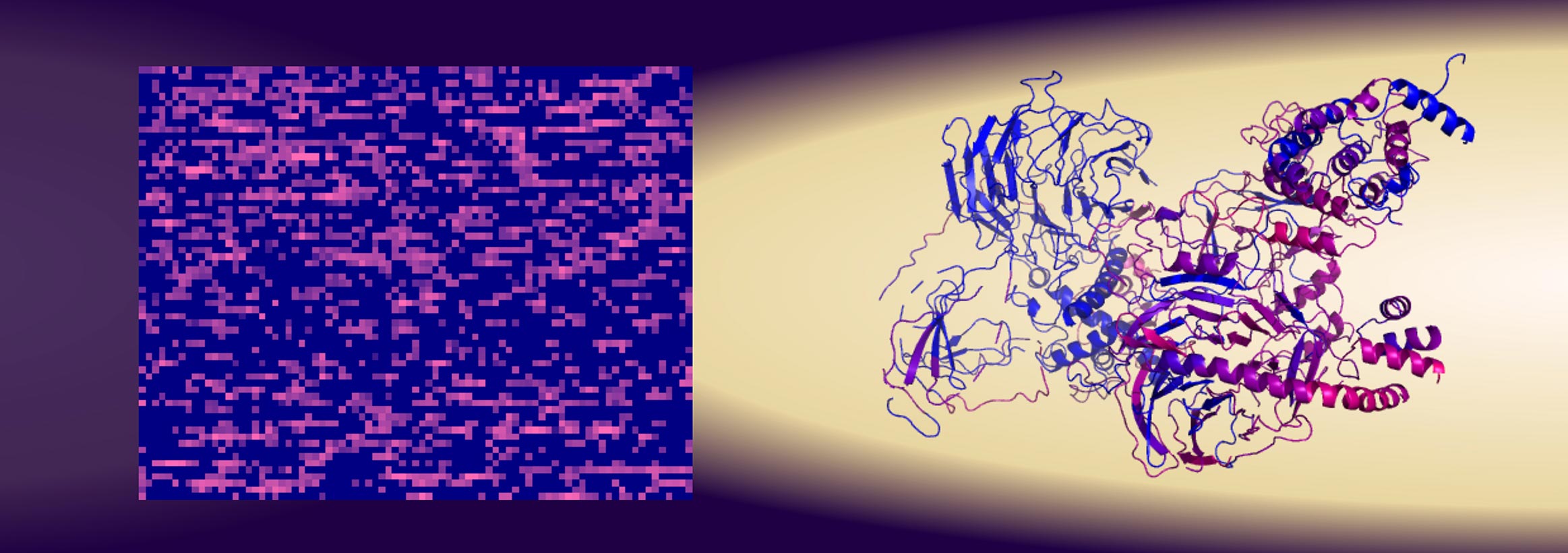
Researchers at the University of Gothenburg have developed a novel method for identifying protein functions without relying on their structures. Traditionally, protein functions were thought to be controlled by their atomic arrangements; however, many proteins lack well-defined structures. Instead, led by Professor Gergely Katona, researchers scanned proteins by assessing the quantity and type of amino acids they contain. This approach revealed that the amino acid content is crucial for binding to the protein survivin, achieving a predictive reliability of about 80%, surpassing traditional methods based on primary structures.
Their experiments identified thousands of 15-amino-acid peptides, confirming that structure had minimal impact on binding strength to survivin. The method, enhanced by machine learning, could expedite the development of new biological drugs. Notably, researchers uncovered a direct link between survivin and another protein, PRC2, which regulates various DNA functions and is associated with cancer. Discovering this connection suggests that cancer drugs targeting survivin and PRC2 may need rethinking to mitigate side effects. Ultimately, the research highlights the potential for identifying new therapeutic targets by focusing on amino acid sequences. The study was published in the journal iScience.


















